|
Home Biography Research Few Publications Full List of Publications Graphical Abstract Project Funding Collaborators Courses Group Members Photo Gallery |
||

|
||
|
|
||
| Just-Mix-&-Read Strategy: Genotyping Single Nucleotide Polymorphism (SNPs) for Personalized Medicine | ||
|
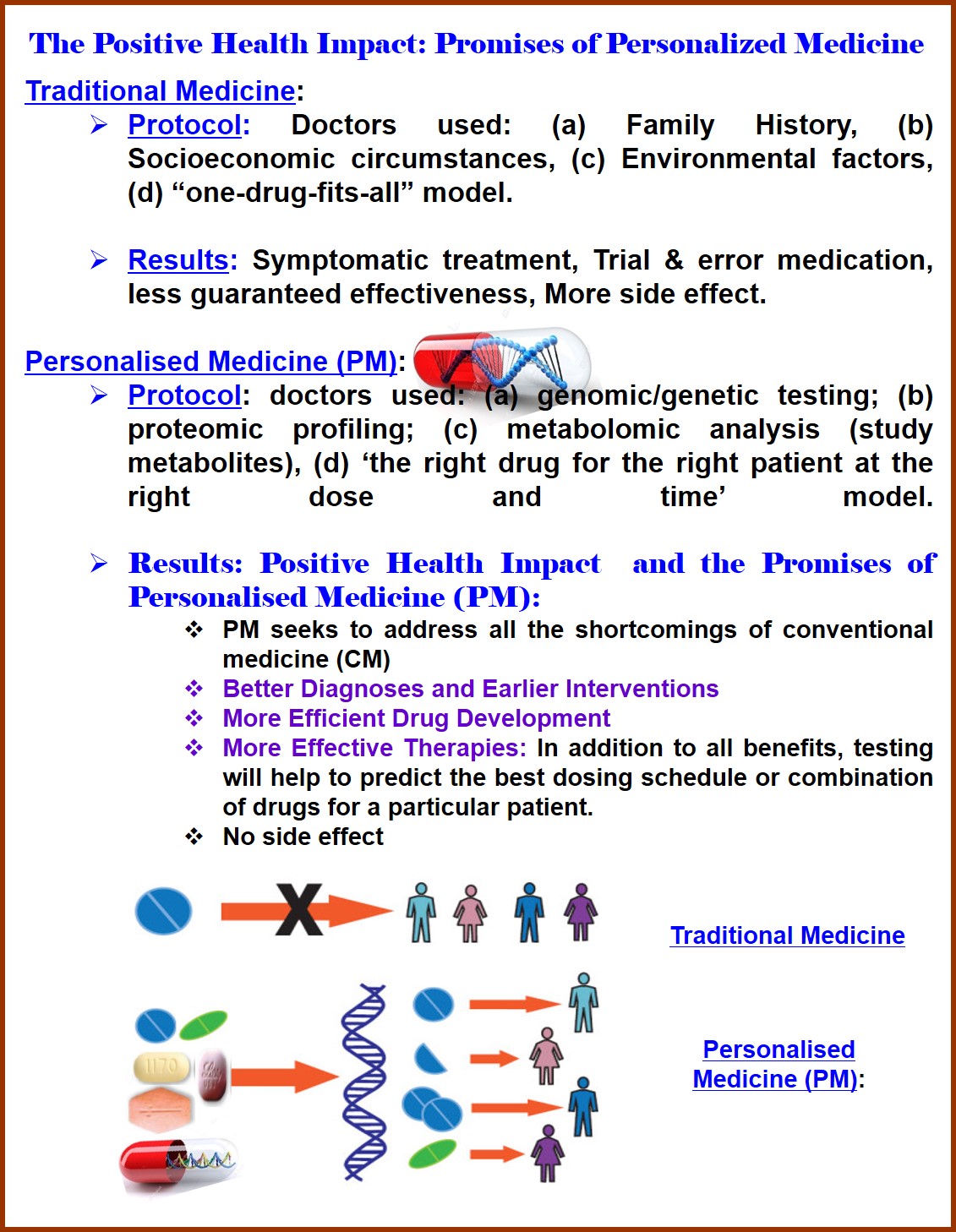
Overview: The mapping of the human genome (Human Genome Project), cataloguing of all the disease related genetic variations/polymorphisms (Hap Map Project), and sharing such information in clinical practice [The Human Variome Project (HVP)] are the major scientific milestones that have opened the door to new approaches to understand the diseases at molecular level and to treat according to genotype going beyond the present day’s traditional “one-drug-fits-all” model medicine (based on phenotype). Mostly the Cancer and cardiovascular diseases are two areas in which genomics are showing promise for treatment advances, although challenges remain. Among all other genetic variations, scientists think Single Nucleotide Polymorphisms (SNPs) are the key enabler to realize the concept of Personalized Medicine, “the right drug for the right patient at the right dose and time”. Scientists see SNPs as a potential tool to improve diseases DIAGNOSIS and TREATMENT planning. They suspect that SNPs may play a role in the different responses to treatments seen among cancer patients and they think that SNPs may also be involved in the different levels of individual cancer risk observed. Thus, genetic variations/SNPs are attractive target for better understanding the genetic basis of complex diseases, and to realize the PERSONALISED MEDICINE. Therefore, Genomic Variations/Biomarkers are the foundation of Personalised Medicine. Personalized medicine hopes to use these genetic variations, especially, SNPs, to develop new safe and effective treatment planning for genetically defined sub-groups of patients. It uses new methods of molecular analysis to better manage a patient’s disease or predisposition toward a particular disease in the context of the patient’s unique genetic and environmental profile. Such approaches may include genetic screening programs that more precisely diagnose diseases and their sub-types, or help physicians select the type and dose of medication best suited to a certain group of patients, thereby minimize or nullifying the side effect. Personalized Medicine is the drugs that cure disease without adverse effects. This new science should reduce the trial-and-error approach to the choice of treatment and thereby limit the exposure of patients to drugs that are not effective or are toxic for them. Therefore, because of their significant biological and clinical importance, recently, there has been a growing interest in single nucleotide polymorphisms (SNPs) typing. In the early days, fluorescent methods of DNA analysis were highly dependent on fluorescent dyes that can complex the duplex DNA by means of non-covalent forces. Then, nucleoside building blocks containing different kinds of fluorophores (covalently linked) were used exclusively. Later on, especially in the post-genomic era, sequence specific complementary binding of short synthetic oligonucleotides containing fluorescent tags have been used in DNA analysis. Today such fluorescent oligonucleotides, called hybridization probes, play a very significant role in qualitative as well as quantitative analysis of the genome and in detection of SNPs. A tremendous progress has been achieved due of intense research efforts, especially in the postgenomic era, by several research groups in designing chemistry, platforms and protocols for DNA detection and SNPs typing, to reach the goal of personalized medicine. Though many methods are being developed for homogenious detection of genetic mutations especially SNPs, it is quintessential to improve the present techniques or to develop new techniques for simultaneous detection of multiple SNPs and copy number variations (CNV) mapping. Also, such a fresh wave of research initiatives should be aimed at devising more costeffective, high throughput, fast and highly accurate analytical methods. It is strongly believed that in due course of time such concurrent technological advancements will lead to new diagnostic and treatment options. Also there is no focus of DNA detection utilizing the unnatural base pairs. With the proposed fluorescent unnatural base pairs, the formation of cross linked DNA via azide alkyne coupling (“CLICK” Chemistry) was not studied till the date. Thus there is a need to develop conceptually new and novel method, and conceptual probe design via new and novel fluorescently labeled base or fluorescent unnatural base for the detection of SNPs and thereby to reach the goal of personalized medicine (J. Photochem. Photobiol. A: Chem. 2020, 388, 112186).
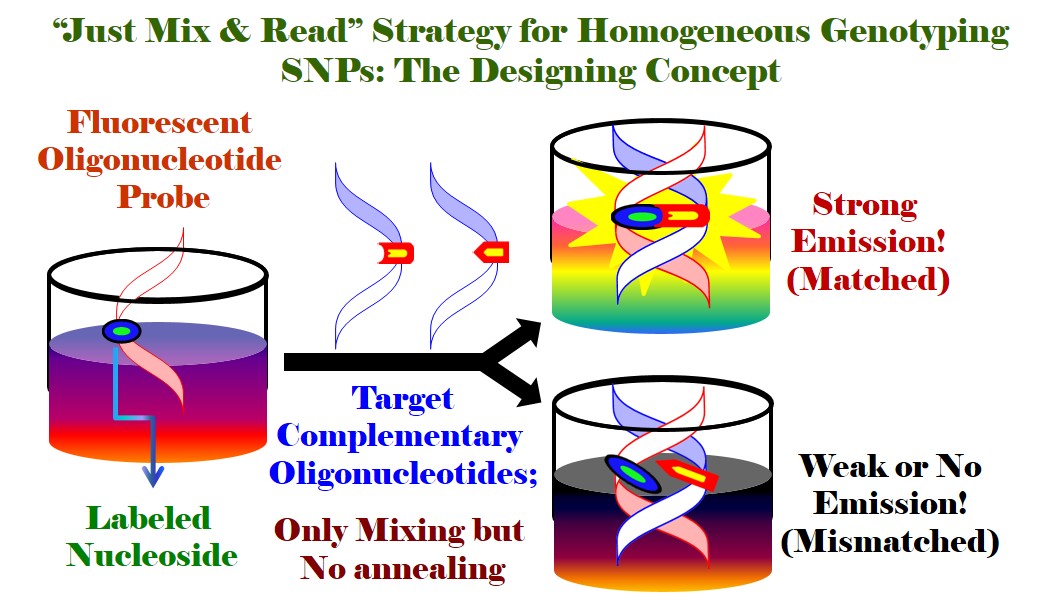
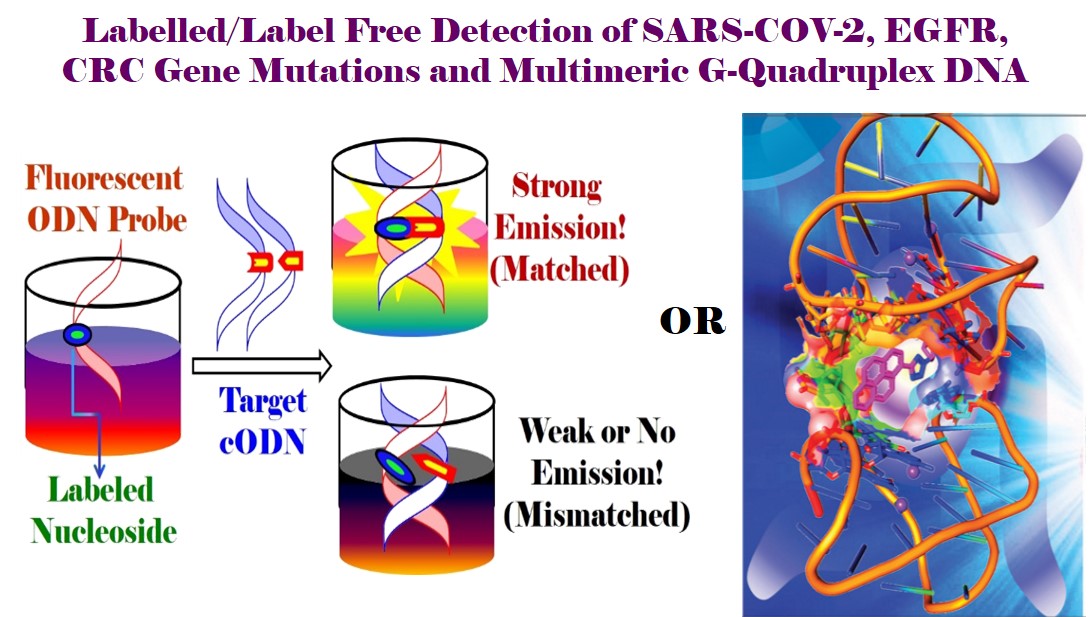
In the field of Single Nucleotide Polymorphisms (SNPs) Genotyping for Personalized Medicine, we have made outstanding contributions. We have fundamentally established “Just-Mix-&-Read Strategy” [Ref. 4, 8, 23, 26, 35, 40, 46] for SNPs genotyping, which is very simple and does not require any stringent hybridization conditions such as DNA annealing [Ref. 1, 3-5, 35, 38-42, 47, 55-57]. The protocol involves Just mixing of two single stranded DNA to offer a stable DNA duplex at room temperature followed by reading the emission signal either emitted from a bare fluorescent probe or from one single stranded fluorescent DNA acting as a probe. The “Just Mix & Read Strategy” is coined, for the first time, in 2013 in Tetrahedron Lett. for the sensing of biomolecules and label-free discrimination of DNA containing a triple T–C/T–G mismatch pair with a fluorescence light-up probe, triazolylpyrene (TNDMBPy) [Ref. 46]. This protocol is further popularized in my subsequent research works for the detection of SNPs and label-free sensing of other DNA lesions [Ref. 3, 4, 8, 23, 26, 35, 40]. This protocol would have potential impact for the practical development of a fluorescent DNA probe system for mutation analysis without labelling a target DNA in a rapid, parallel and cost saving way. Our concept of electronic coupling among an organic chromophore and the π-electron cloud of a nucleobase is highly inspiring for the development of fluorescently labelled nucleobases and oligonucleotide (ODN) probes with novel and modulated photophysical property useable for DNA detection. Thus, I have developed, for the first time, a wavelength shifting fluorescent ODN probe that is found to be efficient in SNPs detection which is the recent target of all biomedical researchers toward reaching the goal of “Personalized Medicine” (Bioorg. Med. Chem. Lett. 2014, 24, 4678). Such a single C–C linked fluorophore labeled solvatochromic nucleoside and fluorescent ODN probe is unique in design. The concept of wavelength shift-guided DNA detection with distinctly enhanced emission intensity makes the ODN probe universally appealing and shed light for the development of long wavelength emissive fluorescent ODN probes for cellular DNA detection. Label-free sensing of DNA lesions is highly important due to their low cost, operational simplicity and fast response times. Utilizing “Just Mix & Read Strategy”, I have also contributed for label-free sensing of DNA lessions such as base mismatches, abasic site and bulge DNA [Ref. 3 8, 23, 26, 46, 47]. Thus, I have reported a novel label-free strategy for the detection of triple T-C mismatched DNA via the discrimination of triple T-G mimasched DNA via the generation of distinct fluorescence signal at visible region utilizing the triazolylpyrene (TNDMBPy) fluorescent probe [Ref. 46]. I have elegantly extended the “Just Mix & Read Strategy” by exploiting his fluorescent unnatural tetrazolylpyrene nucleobase analog (TzPyBDo) as a versatile fluorescent light-up probe for label free detection of pyrimidine base mispair by discriminating the purine base mispair with high sequence specificity. The smart trifunctional probe is also found to detect the abasic site paired against pyrimidine bases with a strongly enhanced fluorescence signal. Furthermore, it represents an attractive probe for recognizing pyridine base bulge-out in DNA bulge hairpin via the generation of enhanced fluorescence signal [Ref 23]. Later on I have further brilliantly exploited the same probe, fluorescent tetrazolyl pyrene nucleoside (TzPyBDo), for specific sensing of dimeric H45 G-quadruplex DNA via a fluorescence light-up response without compromising the conformational stability. The strong binding of the probe via intercalative stacking interaction inside the connecting loop of two G-quadruplex units of H45 and discrimination to other monomeric and long DNA duplexes are accompanied by a drastic enhancement of emission intensity without compromising the conformation and stability. Thus, we utilized tetrazolylpyrene unnatural nucleoside as a human telomeric multimeric G-quadruplex selective switch-on fluorescent sensor (Org. Biomol. Chem. 2017, 15, 10145. Outside Front Cover Page). My thought of progressive utilization of the created molecules lead me to exploit our earlier developed ethanol sensor [Ref. 24] for the detection of DNA lesion. Thus, I have recently shown that pyrenylamido triazolyl aromatic amino acid scaffold (PyAm-ArTAA) probe which was developed for the discrimination of methanol from ethanol is also efficient for the detection of an abasic DNA via aggregation-induced enhanced emission (AIEE) [Ref.8]. We have also synthesized g-oxo-pyrene carboxamide labeled solvatochromic 2'-deoxyuridine (oxo-PyU). Upon incorporation into short oligonucleotides, the synthesized singly- and doubly-labeled fluorescent oligonucleotide probes were found to detect efficiently the opposite base “A” and consecutive “AA” base of a target DNA sequences opposite to the labeled base via generation of enhanced fluorescence signal. We also showed that fluorescence of Oxo-PyU is highly quenched by G-base, thus could be used for designing G-quenched molecular beacon for DNA detection. The probes also could be useful for discrimination of A/G or AA/GG allel as revealed from their fluorescence behavior (Bioorg. Med. Chem. Lett. 2010, 20, 3227). These unique probes can also be utilized for the generation of G-quenched molecular beacon probes for gene sensing. The efficiency of BDF-based technology makes the developed probes universally appealing and might shed light for the design of a probe system for mutation analysis without labelling a target DNA in a rapid, parallel and cost saving way. The detection and targeting of both the mismatched and abasic DNA is highly important which would ultimately help in designing new diagnostics and chemotherapeutics. Furthermore, sensing and targeting the bulge sequence with a fluorescent probe would be useful to study the role of bulges in nucleic acid function or could have significant therapeutic potential. Thus, detection of specific bulges by small fluorescent molecules is an attractive research area since the past several years. Many attempts have been made to prepare such compounds. Towards this end, we reported a label free strategy for the detection of pyrimidine base mismatches (T/T and C/C), sensing of abasic site, and pyrimidine base bulge DNA using an unnatural tetrazolylpyrene nucleoside (TzPyBDo) as a bare fluorescent probe. The H-bonding/hydrophobic force mediated interactions allow the sensing of all three deformed DNA via an enhancement of fluorescence signal using our simple “Just-Mix and Read” strategy. The binding of the probe to all the three deformed DNA duplexes is accompanied by an increase in the thermal melting stability of the deformed DNA. That the probe binds efficiently to the minor groove near the deformed site was evident from spectroscopic study. All the spectral evidences open up a multitude of possibilities for using our probe, tetrazolylpyrene nucleoside, as an efficient fluorescent light-up bio-probe for label free DNA detection (J. Photochem. Photobiol. B 2017, 173, 165-169). In the same line earlier we published, Unnatural triazolyl nucleoside stabilizes an abasic site containing DNA duplex equally as the stabilization of a natural A-T pair. We also achieved better than reported abasic duplex stabilization with novel triazolylphenanthrene nucleoside TPhenBDo. The large surface area, polarizability and strong stacking propensity of triazolylphenanthrene play a major role to offer thermal stabilization of TPhenBDo-F duplex comparable to that of a natural A-T pair via strong intercalative stacking interaction (RSC Advances 2013, 3, 21352). A novel label free strategy was adopted to detect DNA base mismatch via the generation of distinct fluorescence signal at visible region utilizing the same probe. Also binding to the minor groove of calf-thymus DNA (ct-DNA) and strong binding in hydrophobic pocket of bovine serum albumin (BSA), switched-on the fluorescence of smart fluorescent probe, triazolylpyrene (TNDMBPy) (Tetrahedron Lett. 2013, 54, 2627-2632). Forster resonance energy transfer (FRET) is a highly efficient strategy in illuminating the structures, structural changes and dynamics of DNA, proteins and other biomolecules and thus is being widely utilized in studying such phenomena, in designing molecular/biomolecular probes for monitoring the hybridization event of two single stranded DNA to form duplex, in gene detection and in many other sensory applications in chemistry, biology and material sciences. Moreover, FRET can give information about the positional status of chromophores within the associated biomolecules with much more accuracy than other methods can yield. Toward this end, we reported the ability of fluorescent unnatural nucleoside, triazolylphenanthrene (TPhenBDo) to show FRET interaction upon hybridization with fluorescently labeled natural nucleosides, PerU or OxoPyU forming two stable chimeric DNA duplexes. The pairing selectivity and the thermal duplex stability of the chimeric duplexes are higher than any of the duplexes with natural nucleoside formed. The hybridization results in a Forster resonance energy transfer (FRET) from donor triazolylphenanthrene of TPhenBDo to acceptor oxopyrene of OxoPyU and/or to perylene chromophore of PerU, respectively, in two chimeric DNA duplexes. Therefore, we have established the FRET process in two chimeric DNA duplexes wherein a fluorescently labeled natural nucleoside (OxoPyU or PerU) paired against an unnatural nucleoside (TPhenBDo) without sacrificing the duplex stability and B-DNA conformation. The hybridization accompanying FRET event in these classes of interacting fluorophores is new. Moreover, there is no report of such designed system of chimeric DNA duplex. Our observed phenomenon and the design can potentially be exploited in designing more of such efficient FRET pairs for useful application in the detection and analysis of biomolecular interactions and in material science application (J. Photochem. Photobiol. B: Biology 2016, 162, 669-673). We recently published Design of “Click” Fluorescent Labelled 2′-deoxyuridines via C5-{4-(2-Propynyl(methyl)amino)}phenyl acetylene as a Universal Linker: Synthesis, Photophysical Property and Interaction with BSA.In this particular, we report the rational design and synthesis of triazolyl push-pull fluorophore-labeled uridines via the intermediacy of C5-[4-(2-propynyl(methyl)amino)]phenyl acetylene as a universal linker. The synthesized nucleosides showed interesting solvatochromic characteristic and/or intramolecular charge transfer (ICT) features. A few of them also exhibited dual-emitting characteristics evidencing our designing concept. The synthesized triazolyl benzonitrile (10C), naphthyl (10E), and pyrenyl (10G) nucleosides were found to exhibit interesting ICT and dual (LE/ICT) emission properties. The dual-emitting pyrenyl nucleoside maintained a good ratiometric response in the BSA protein microenvironment, enabling the switch-on ratiometric sensing of BSA as the only protein biomolecule. Thus, it is expected that the new fluorescent nucleoside analogues would be useful in designing DNA probes for nucleic acid analysis or studying DNA-protein interactions via a drastic change in fluorescence response due to a change in micropolarity (J. Org. Chem. 2018, 83, 7606–7621; John Wiley & Sons (2016). (ISBN: 978-1-118-17586-6); Elsevier, 2018 (ISBN: 9780128112922). Currently, we are exploring a conceptually new array design to immobilize fluorescent oligonucleotide probes for detecting the particular un-labelled target gene sequence. Our system expectedly would be able to perform high-throughput mutation analysis and disease diagnosis in a parallel, cost-saving and label-free way.
In summary, we have developed several conceptual probes/techniques and were able to use them for gene detection successfully. In this way development of simple technology might able to help in detection of all the SNPs of Human genome. Thus, ongoing, novel, easy and high throughput detection techniques, microarray, expectedly would allow to us detect SNPs in large scale and thereby we will be able to make SNPs profile of a person. This will allow physicians to compare with the global mutations repository and to diagnose a particular disease associated with a mutation and to identifying genetic susceptibility to disease and to study pharmacogenomics to revolutionize the practice of medicine by individualisation of treatment through the use of novel diagnostic tools. Finally, in this way, pharmacogenomics would reduce the trial-and-error approach of treatment and thereby limit the exposure of patients to drugs that are not effective or are toxic for them or have serious, side effects. Thus, our continuous efforts blended with pharmacogenomics would allow us, one day, to fix the disease at the Genetic level and to provide tailor medical treatment (Personalised Medicine) to the individuals with all round positive health impact. As a continuous effort of our drug design and development project along with study of Drug -DNA/Drug-Protein interaction and the above knowledge would help in designing genetically effective Personalised Drugs. The better understanding of genetic variations could help scientists to identify new disease subgroups or their associated molecular pathways. Thus, it would help in designing drugs that target those subgroups. Molecular analysis could also help to select patients for inclusion in/ exclusion from the late stage clinical trials which would help to approve drugs that might otherwise be abandoned because they appear to be ineffective in the larger patient population. Therefore, our present fundamental and applied contributions will have potential impacts to the strengthening of both basic and applied health research not only in the country but also to the world scenario especially in reaching the Goal for Personalised Medicine (PM)-the concept of treating a person with “the right drug for the right patient at the right dose and time” model to overcome all the shortcomings of conventional medicine (CM)/traditional medicine (TM)-thus, to give benefit to the all Human Civilization worldwide.
|
||