|
Home Biography Research Few Publications Full List of Publications Graphical Abstract Project Funding Collaborators Courses Group Members Photo Gallery |

|
|
|
| EXPANSION OF THE GENETIC ALPHABETS |
|
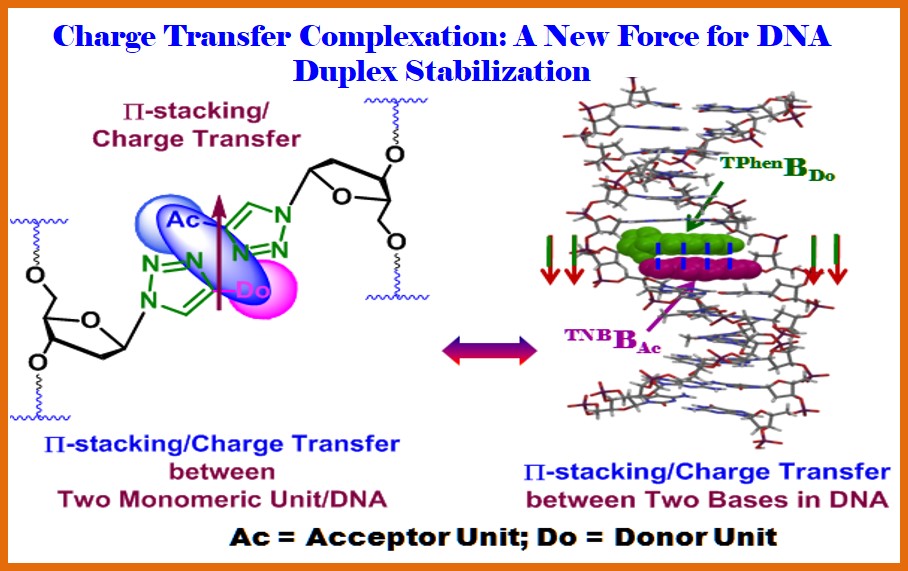
Overview: Our pioneering work for Expansion of the Genetic Alphabet includes unraveling the Charge Transfer (CT) Complexation as a strong enough force to stabilize a DNA Duplex. Specific pairing of nucleobases is the basis of genetic alphabet, ultimately leading to the basis of genetic code. An expanded genetic alphabet would enlarge the informational and functional potential of DNA for both in vitro and in vivo applications (J. Nucleic Acids 2012, Article ID 718582). Search for a new and biologically significant “interacting force” in base pair stabilization, in a duplex DNA, except those reported, has led us to think charge transfer (CT) complexation force and to design triazolyl donor-acceptor base pair(s). Toward this end the following innovations have come up with immediate/long term applications. Toward this end we have unraveled that the CT complexation force between unnatural triazolyl donor-acceptor nucleoside pair is strong enough to stabilize a DNA duplex efficiently which could efficiently be replicated (J.Org.Chem. 2013, 78, 278; Curr. Protoc. Nucleic Acid Chem. 2015, 58:1.32.1-1.32.27; Org. Biomol. Chem. 2016, 14, 5088). Thus, the concept of non-H-bonding unnatural nucleobase surrogates and the power of charge transfer (CT) interaction force are unified in my donor-acceptor unnatural triazolyl nucleobases synthesized via the novel click approach. Such a donor and an acceptor nucleobase pair (TPhenBDo : TNBBAc) in DNA is found to show excellent pairing selectivity via charge transfer interaction in any sequence context. Further, the new and novel triazolyl unnatural donor-acceptor nucleobase pair offer a good stabilization of the hetero-duplex that is comparable to that of a natural A/T pair. The CT interaction force could well be recognized by DNA polymerase to copy DNA containing such a donor-acceptor triazolyl unnatural base pair [Ref. 1, 5, 10, 14, 22-23, 26, 30-31, 41-42, 47, 50]. The unique and innovative idea and the exploration of unnatural hetero-duplex stabilization via charge transfer interaction are universally appealing and might open a new avenue in the field of expansion of the genetic alphabet and in modern synthetic biology.
We have also exploited the same probe in stabilizing an abasic DNA lesion (RSC Adv. 2013, 3, 21352). Targeting an abasic site with designed nucleobase analogs, DNA probe, or small molecules is a promising route in the development of therapeutic and diagnostic agents directed toward base excision repair (BER)-deficient cancers. To this end, I have exploited the DNA probe containing unnatural triazolylphenanthrene nucleoside TPhenBDo and shown that it stabilizes an abasic DNA lesion in a better way than any of the reported nucleosidic/nonnucleosidic base analogs [Ref. 47]. The excellent stabilization makes the probe universally appealing and might shed light for the design of long wavelength emissive fluorescent sensors and chemotherapeutics targeting the abasic DNA [Fluorescent Analogues of Biomolecular Building Blocks: Design and Applications, Eds. Tor, Yitzhak; Wilhelmsson, Marcus. Publisher: John Wiley & Sons (2016). (ISBN: 978-1-118-17586-6).; Sonogashira Cross-Coupling: Alkyne-modified Nucleosides and their Applications, In Palladium-Catalyzed Modification of Nucleosides, Nucleotides and Oligonucleotides, Elsevier (2018, ISBN 9780128112922, Book Chapter)]; (Tetrahedron, 2019, 75, 3024]. In continuation of our earlier research towards expansion of genetic alphabets, we recently reported the design of Fused Triazolyl Phenanthrene! as a new generation of doubly widened hydrophobic unnatural nucleobase surrogate having interesting photphysical property with chrysene type emission. Theoretical study based on Molecular dynamics simulation using Schrodinger Macromodel and DFT reveal its potential stacking propensity in a duplex DNA without compromising the right-handed B-form helical conformations with C2'-endo/C3'-exo sugar disposition. Thus, the novel FTPhen is expected to find wide applications in chemistry, biotechnology and DNA-based material design (Chemistry Select 2017, 2, 3577-3583.). We also recently reported the design and synthesis of a novel fused triazolyl2-quinolinone (FTQuon) nucleoside as a new generation of angularly widened unnatural nucleobase surrogate with two possible H-bonding faces-one H-bond acceptor and another donor. The synthesis via a tandem CuAAC-Ullmann coupling, the study of photophysical properties and theoretical study in the context of DNA are the main contents of this report. The newly designed nucleoside shows interesting photphysical property with slight blue shifted solvatochromicity. It also shows pH sensitive emission. All the theoretical DNA duplexes containing the FTQuon show right-handed B-form helicity as revealed from a molecular dynamics simulation using Schrodinger Macromodel. A theoretical (DFT) study indicates a good stabilizing property of FTQuon via pairing with natural pyrimidine bases. It also shows good interaction property with BSA protein signalled via a switch on fluorescence response (Tetrahedron 2018, 74, 2218). Both the nucleobases are unique contributions in the expansion of the genetic alphabet [Ref. 50]. These nucleobases also showed antibacterial effects and are under consideration for testing antiviral activity against HIV and SARS-CoV-2. The Merck Lifescience and Rasayan Inc., USA has come forward for collaborative work for the design and development of DNA-based material [Ref. 14, 22]. We also synthesized Triazolyl C-Nucleosides via the Intermediacy of β-1'-ethynyl-2'-deoxyriboside Derived from a Nicholas Reaction (Tetrahedron. 2019, 75, 3024). Among many other contributions, two more classes of nucleobases are under consideration by Sapala Organic Pvt. Ltd., Hyderabad, for large scale production and a detailed study for antiviral activity. These are: (i) Triazolyl C-Nucleobase analogs via the Intermediacy of β-1'-ethynyl-2'-deoxyriboside derived from a Nicholas Reaction and (ii) donor/acceptor aromatics decorated tetrazolyl nucleobase analogs via a stereospecific and regioselective SN2 pathway [Ref. 10, 30]. We recently have explored the stereospecific and regioselective synthesis of donor/acceptor tetrazolyl nucleosides (Bioorg. Med. Chem. Lett. 2016, 26, 2044).
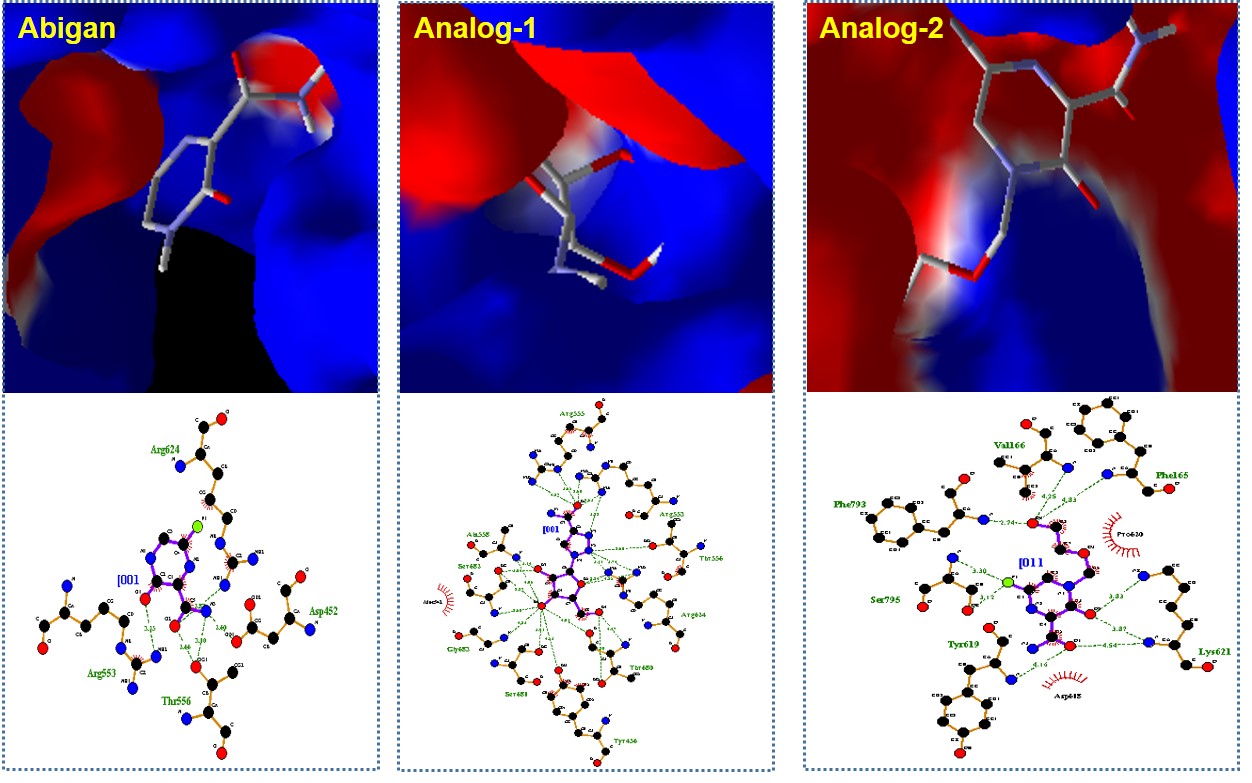
Application-Nucleoside Based Drug Design: As a part of nucleoside based drug development research, I have excellently exploited a fluorescent nucleoside, tetrazolylpyrene unnatural nucleoside (TzPyBDo), as a probe for targeting and sensing of H45 multimeric G-quadruplex DNA. The uniqueness of this work is evident by its appearance in Outside Front Cover Page of Org. Biomol. Chem. in 2017 (Org. Biomol. Chem., 2017, 15, 10145, Front Cover Page) and sensing of protein BSA (Communicated). Furthermore, the excellency and the specificity of the probe is also exploited for label-free sensing of pyrimidine base mismatches (T/T and C/C), abasic DNA and bulge DNA (J. Photochem. Photobiol. B 2017, 173, 165). All these findings are evidencing to open up a multitude of possibilities for using this nucleoside as an efficient fluorescent light-up bio-probe for label-free gene detection. A preliminary study has shown that the nucleoside is potent against the Colorectal cancer cell line. Details study is undergoing. The developed unnatural oligonucleotides [Ref. 35, 38-42, 47, 55, 57] might find potential applications in “Antisense Gene Therapy,” in particular for the treatment of Neurological Disorder and Inflammatory Bowel Disease. We have currently started a project for the synthesis of Abigan (favipiravir) analog, a potent drug against COVID-19. SARS-CoV-2 (2019-nCoV) coronavirus is posing dangers worldwide at the very moment. Currently, the clinical treatment of the disease, COVID-19, is mainly symptomatic combined with the repurposing of already marketed antiviral drugs such as ritonavir/lopinavir, remdesivir, a combination of ritonavir/lopinavir with interferon beta, and antibiotics such as Chloroquine and hydroxychloroquine to treat secondary infections. Therefore, there remains an urgent need and challenge to save human life worldwide by developing specific antiviral therapeutics and vaccines against SARS-CoV-2. RNA-dependent RNA polymerase (RdRp) plays crucial role to make multiple copies of viral genomic RNA. Thus, design of inhibitor drugs for RdRp is an excellent strategy to stop viral replication and hence spread of viral diseases. Recently, it has been found that some of the drugs, such as, ribavirin and remdesivir, developed for SARS, MERS etc. are also effective for SARS-CoV-2. The approved influenza drug, Favipiravir (Avigan) selectively inhibits the RNA polymerase of the influenza virus, an enzyme required for viral replication once human host cells are infected. COVID-19 also uses this enzyme to replicate and is classified into the same type of single-stranded RNA virus like influenza. Recently, Abigan is used as the only drug to help tackle the spread of the COVID-19 pandemic in Japan. Phase III clinical trials are ongoing in Japan. In the US, Fujifilm started Phase II trials earlier in April 2020. The structures of Avigan and other antiviral drugs which are being used for COVID-19 treatment and the target analogs are given below. In this Global crisis and as a continuation of the research efforts for the synthesis of novel nucleosides, I recently involved in developing antiviral nucleosides such as Abigan and AICAR analogs. Considering the effectiveness of Abigan against Coronavirus and the side effects of other nucleoside based drugs, we came up with few target designed analogs.
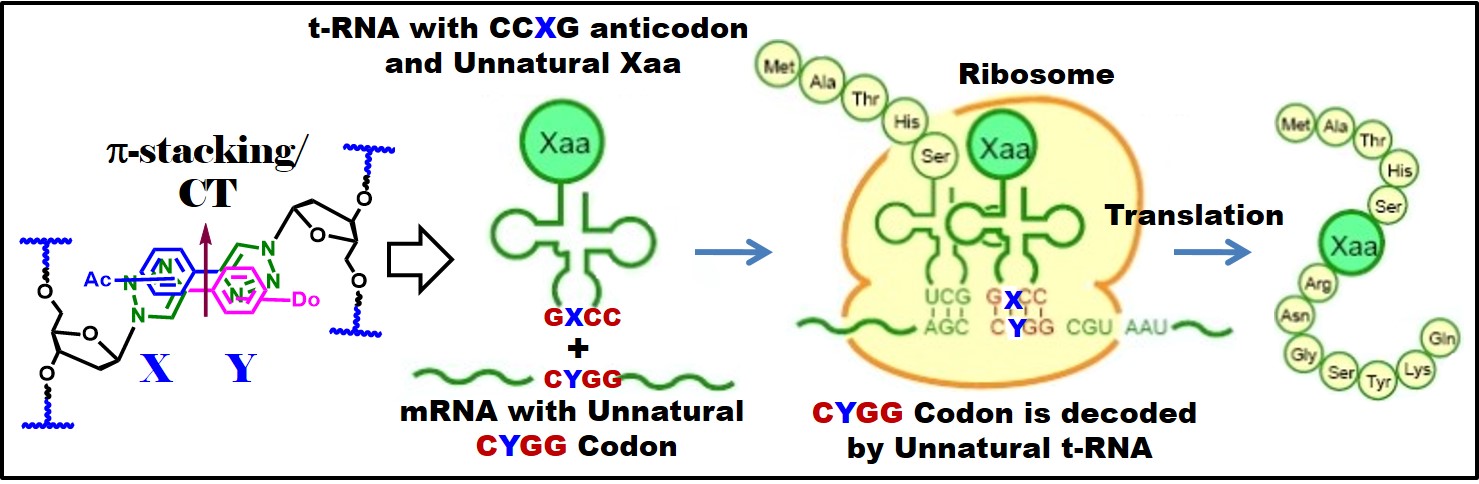
Our preliminary Molecular Docking study suggested them replacing the ribose moiety by linear alcohol as is there in acyclovir. Furthermore, the triazole containing bases shows strong interaction and binding effect reflecting their activity to stop the extension of RNA of the viruses. The target nucleosides are believed to have mechanism of action which is similar to Abigan. This work would yield a highly effective COVID-19 drug. Application-DNA Scaffolds in Photochemistry and Photobiology: I have extended my fundamental contributions in the field of photochemistry and photobiology utilizing the developed fluorescent DNA probes. Thus, I have reported the hybridization accompanying Förster Resonance Energy Transfer (FRET) process in chimeric DNA duplexes containing unnatural nucleoside, triazolylphenanthrene (TPhenBDo) and labeled natural nucleosides, PerU or OxoPyU [J. Photochem. Photobiol. B: Biology 2016, 162, 669]. The hybridization accompanying FRET event in these classes of interacting fluorophores is new. Moreover, there is no report of such designed system of chimeric DNA duplex. The concept can potentially be exploited for useful application in the detection and analysis of biomolecular interactions and material science applications. We also presented the serendipitous discovery of what we call a Dual Door Entry! to exciplex formation, occurring in a chimeric DNA duplex containing a non-nucleoside-nucleoside pair. The nucleobases packed inside the duplex via intercalative stacking interaction leading to exciplex emission either via FRET from donor unnatural nucleoside, TPhenBDo or direct excitation of FRET acceptor unnatural non-nucleosidic base surrogate, OxoPyS. To the best of our knowledge, the design of chimeric DNA duplex wherein a fluorescent nucleosidic base surrogate paired against a non-nucleosidic base surrogate involved in distance dependent energy transfer process or exciplex formation is not known. Moreover, oxopyrene fluorophore as FRET acceptor is new. This is the start of a new generation of probes which could find wide applications in the field of chemical biology and in designing light harvesting DNA materials. This novel and important finding that will further define research to establish and to create more of such conceptually designed DNA system for future application in optoelectronics, chemical biology, and in material sciences. This study will surely draw attention of broad scientific community. (Chem. Commun. 2014, 50, 829). This unique photophysics of Dual Door Entry has been extended and proved to be equally applicable to many other nucleic acids/peptide/β-lactam-based designs [Ref.12, 34]. The discovery has embarked on the uniqueness and universality of the Dual Door Eentry system, which could find wide applications in the field of chemical biology and in designing light-harvesting DNA materials. This system has attracted the attentions of various scientists working in the field. This finding would open up a new field in photobiology. Our long term goal is to use the efficient new unnatural nucleoside base pair/pairs to drive the synthesis of unnatural proteins containing our designed unnatural amino acids. We have a hope to translate an expanded genetic alphabet into an expanded genetic code. Thus, a combination of both the project will help in translating the developed expanded genetic alphabet to an expanded genetic code which provocatively, may even lead to the assembly of such a system within a living cell, potentially creating a semi-synthetic organism and life with increased diversity. The unnatural oligonucleotides might find potential applications in “Antisense Gene Therapy”. The new nucleoside base analogues that will be obtained may have potential biological activity and may find clinical use in future for the treatment of viral diseases such as AIDS, cancer etc. |