Welcome to Aniket's Website
PMRF Fellow, Member of the Royal Society of Biology (MRSB), London, NEWGEN IEDC Fellow, Centre for Environment, IIT Guwahati
Prof. Subhendu Sekhar Bag's Laboratory
About me

Aniket Banerjee is a Ph.D. student in the Centre for Environment, at Indian Institute of Technology (IIT) Guwahati, India. He received his undergraduate and post graduate degree in Biotechnology from the School of Biotechnology, Kalinga Institute of Industrial Technology (KIIT), Bhubaneswar, Odisha in 2019. During his M.Tech dissertation he was introduced to two new terms; Expansion of Genetic Alphabet and Expansion of Genetic Code, by Prof. Subhendu Sekhar Bag at IIT Guwahati, which immediately caught his interest. Intrigued by the prospect of designing and developing synthetic genetic materials as well as synthetic proteins with hypothetically any kind of function made him feel passionate towards research which ultimately led him to enrol in a Ph.D. on his favourite field of study. Aniket is a recipient of the prestigious Prime Minister's Research Fellowship (PMRF), awarded to the country's excellent doctoral candidates for pursuing their research.
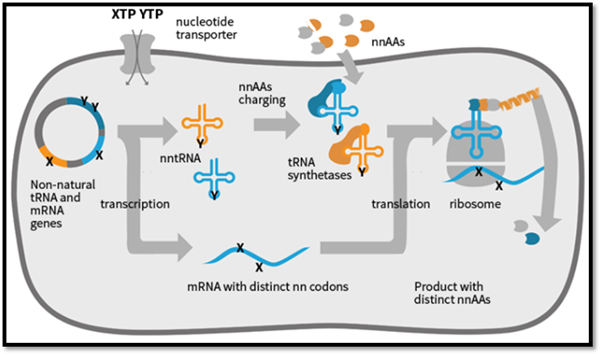
Aniket’s research interests include Expansion of Genetic Alphabet, which means incorporation of unnatural and synthetic base pairs into DNA strands, and the Expansion and Reprogramming of Genetic Code, which means the incorporation of unnatural amino acids into peptide and protein sequences by biological cell free and cell-dependent strategies
Professional Memberships:
1) Member of the Royal Society of Biology (MRSB), London UK.
2) Member of the American Society of Microbiology, USA.
CV 
Supervisor

Prof. Subhendu Sekhar Bag (CChem, FRSC, FICS)
Professor, Department of Chemistry, IIT Guwahati.
Research Area: Bio-Organic/Medicinal Chemistry of Nucleic Acids, Peptides, and b-Lactam Antibiotics.
Phone: +91-361-2582324||Email: ssbag75@iitg.ac.in
Projects
.png)
Expansion of Genetic Code
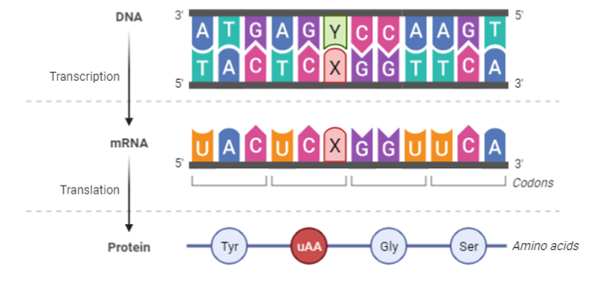
An approach towards utilization of an expanded genetic alphabet for encoding an unnatural amino acid
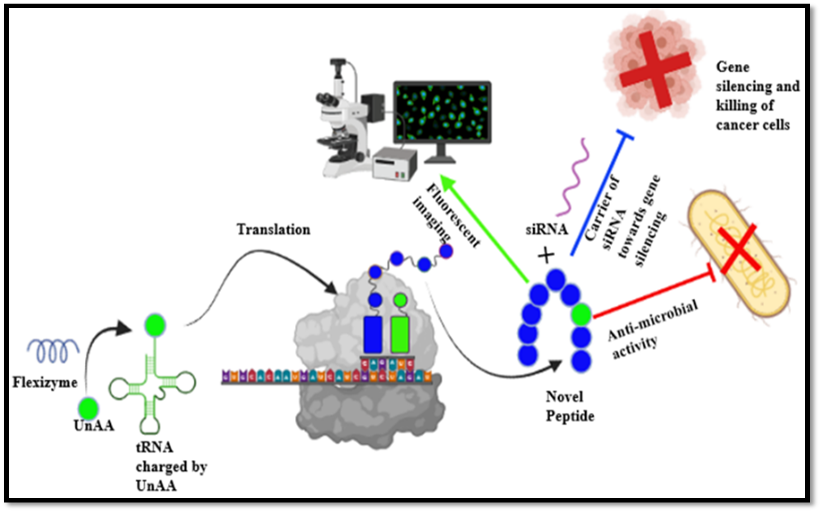
A flexizyme mediated method of reprogramming of genetic code
Work
Expansion of Genetic Alphabet
Motivated by the requirements of labelled RNAs and amino acids, studies revolving around the formulation of alternative base pairs capable of expansion of the genetic alphabet is slowly grabbing the attention of scientists in recent times. The living organisms encode the information stored in the genome, and this genome is made up of a two base pairs, namely A-T and G-C. An expanded genetic alphabet would magnify the informational and functional potential of DNA for both in vitro and in vivo applications by several folds. In order to form a successful base pair, the unnatural bases formed must follow certain rules such as:
- They must maintain stability of the duplex.
- Must be orthogonal to the naturally occurring bases (A, T, G and C).
- Must be recognized and by different polymerases (each of which is highly specific) in order to promote replication, transcription and translation of the unnatural bases.
- The selectivity of incorporation of two unnatural bases must be significantly higher than the selectivity of the individual unnatural bases against its natural counterparts.
Our long term goal is to use the efficient new unnatural nucleoside base pair/pairs to drive the synthesis of unnatural proteins containing our designed unnatural amino acids. We have a hope to translate an expanded genetic alphabet into an expanded genetic code. Thus, a combination of both the project will help in translating the developed expanded genetic alphabet to an expanded genetic code which provocatively, may even lead to the assembly of such a system within a living cell, potentially creating a semi-synthetic organism and life with increased diversity. The unnatural oligonucleotides might find potential applications in Antisense Gene Therapy. The new nucleoside base analogues that will be obtained may have potential biological activity and may find clinical use in future for the treatment of viral diseases such as AIDS, cancer etc.
Expansion of Genetic Code
Barring a few exceptions, the naturally occurring amino acids, i.e the canonical amino acids are generally preserved and encoded in all three kingdoms of life. While, all the natural proteins are formed by the 20 naturally occurring amino acids, the ability to incorporate non canonical amino acids into proteins having specific functionalities are capable of virtually opening up an avenue towards the discovery and further synthesis of new proteins and peptides possessing novel functions, such as therapeutic or diagnostic properties.
Biosynthetic method for incorporation of UAAs:
The biosynthetic method must possess two components:
- Unique codon with its corresponding tRNA pair.
- Aminoacyl tRNA synthetase (aaRS), which is unique for each unnatural amino acid.
The design and encoding of fluorescent unnatural amino acids toward expanding the genetic code would expand proteins functions and biotechnological applications. Our observation on the installation/modulation of emission response via click reaction has led us to design triazolyl donor-acceptor unnatural amino acids with novel solvatochromic fluorescence properties. The conceptual development of fluorescent triazolyl donor-acceptor chromophore decorated unnatural amino acids is the unique introduction to the expanded genetic code. My research work focuses on the expansion of genetic code in a cell-free as well as a cell-based approach
Education
- Indian Institute of Technology, Guwahati (IITG)
- Centre for the Environment.
- Coursework: Completed (9.5 CPI).
-
B.Tech/ M.Tech Dual Degree
- School of Biotechnology, KIIT.
- CGPA: 9.07
Publications
1) Simultaneous sensing and photocatalytic degradation to free water from toxic dye pollutants for possible agricultural and household applications: Role of generated holes and hydroxyl radicals. Subhendu Sekhar Bag*, Sayantan Sinha, Banerjee, Aniket, Animes Golder. Journal of Environmental Chemical Engineering, 2022, 10(6), 108919 https://doi.org/10.1016/j.jece.2022.108919. (IF=7.968)
Conferences and Workshops
1) Indian Science Congress Association: Bhubaneswar Chapter - National Seminar on science and technology for indigenous development in India. (December 9-11, 2015). |
Teaching Experience
Teaching Assistant at North Guwahati College.
Courses taught:
1) Principles of Genetics. Code: ZOO-HC-5026.
2) Molecular Biology. Code: ZOO-HC-5016.
3) Animal Physiology: Life Sustaining Systems. Code: ZOO-HC-4026
Teaching Assistant at IIT Guwahati
Courses taught:
1) Bio-Organic Chemistry. Code: CH 621.