A better version of creating the psf file has been discussed here. Please avoid this page and follow that process.
Pdb files contain only the atom coordinates (and some citation references). However, to apply the bonded interactions in MD simulation, we need to have the information regarding bonds, angles, dihedrals and impropers. This information is contained in PSF files, they also contain the charges to apply the electrostatic interactions. We will generate psf file using Vega ZZ (another open source software, we don't wanna pay for trifle things, right?).
A side note here, though it might not be required for our case, we will create a wide pdb file. When we convert pdb to psf, vegazz will use a distance criteria to determine which atom is connected to which other atom. This means that if packmol packed two ethanol molecules too closely (we specified a tolerance of 2 A, so we are safe) then VegaZZ will connect the two molecules and portray it as one. Computers are egoistic and don't care about what to do, they just do what they can. It can sometimes cause error. To avoid this, people create another pdb file with really large box dimenions. This ensures that no two molecules are too close. As such there won't be any "Bond Order too high" error in VegaZZ. (Be very cautious every time you see this warning in VegaZZ). Follow these steps to create the wide pdb file:
Follow these steps:
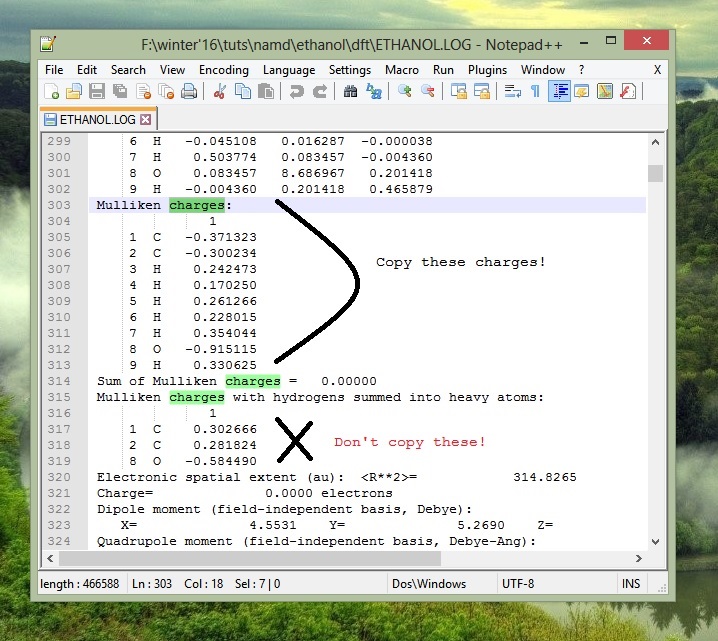
Have a look at the psf file (open it in Notepad++). Do you notice the plethora of question marks in the file? Actually, they are meant to specify the charge of the respective atoms. We obtained the charges from Gaussian calculations (in case you remember), we need to copy those charges in a separate file (in case you haven't till now).
Charges from Gaussian09: (copy ESP charges if you specified the pop=chelpg keyword!)

Next we need to insert these charges into the psf file. Also, we will also give an atomtype to each of these atoms. Why? We are trying to think a step ahead. Our next step will be creating the param file or the force field file. At times it occurs that many different atoms are quiet similar to each other and as such they can be specified the same atomtype. This will save computational time and energy. As such we will give the same atomtypes to atoms that have the same kind of parameters. How do we know if two atoms have the same parameters? Experience helps. Also, you can check the forcefield file first, then edit this file and run it again. For now, just follow these steps:
Previous Page Main Page Next Page